Purdue University researchers are using deep learning to design a novel approach to better understand how proteins interact in the body. This allows them to produce accurate structural models of protein interactions involved in various diseases. This helps to design better drugs that specifically target protein interactions. The work is released online in Bioinformatics.A typical body has more than 20,000 different types of proteins, each of which is involved in many functions essential to human life.
“To understand molecular mechanisms of functions of protein complexes, biologists have been using experimental methods such as X-rays and microscopes, but they are time- and resource-intensive efforts,” said Daisuke Kihara, a professor of biological sciences and computer science in Purdue’s College of Science, who leads the research team. “Bioinformatics researchers in our lab and other institutions have been developing computational methods for modeling protein complexes. One big challenge is that a computational method usually generates thousands of models, and choosing the correct one or ranking the models can be difficult.”
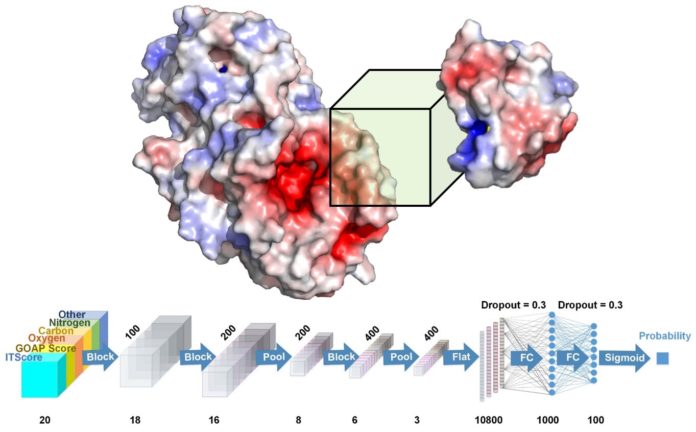
Kihara and his team developed DOVE, DOcking decoy selection with Voxel-based deep neural nEtwork, which applies deep learning principles to virtual models of protein interactions. DOVE scans the protein-protein interface of a model and then uses deep learning model principles to distinguish and capture structural features of correct and incorrect models.
“Our work represents a major advancement in the field of bioinformatics,” said Xiao Wang, a graduate student and member of the research team. “This may be the first time researchers have successfully used deep learning and 3D features to quickly understand the effectiveness of certain protein models. Then, this information can be used in the creation of targeted drugs to block certain protein-protein interactions.”